“A three-dimensional engineered tumour for spatial snapshot analysis of cell metabolism and phenotype in hypoxic gradients” Nature Materials
Darren Rodenhizer, Edoardo Gaude, Dan Cojocari, Radhakrishnan Mahadevan, Christian Frezza, Bradly G. Wouters & Alison P. McGuigan
Nature Materials (2015)
Full Text on ResearchGate
Abstract
The profound metabolic reprogramming that occurs in cancer cells has been investigated primarily in two-dimensional cell cultures, which fail to recapitulate spatial aspects of cell-to-cell interactions as well as tissue gradients present in three-dimensional tumours. Here, we describe an engineered model to assemble three-dimensional tumours by rolling a scaffold–tumour composite strip. By unrolling the strip, the model can be rapidly disassembled for snapshot analysis, allowing spatial mapping of cell metabolism in concert with cell phenotype. We also show that the establishment of oxygen gradients within samples that are shaped by oxygen-dependent signalling pathways, as well as the consequential variations in cell growth, response to hypoxic gradients extending from normoxia to severe hypoxia, and therapy responsiveness, are consistent with those of tumours in vivo. Moreover, by using liquid chromatography tandem mass spectrometry, we mapped cellular metabolism and identified spatially defined metabolic signatures of cancer cells to reveal both known and novel metabolic responses to hypoxia.
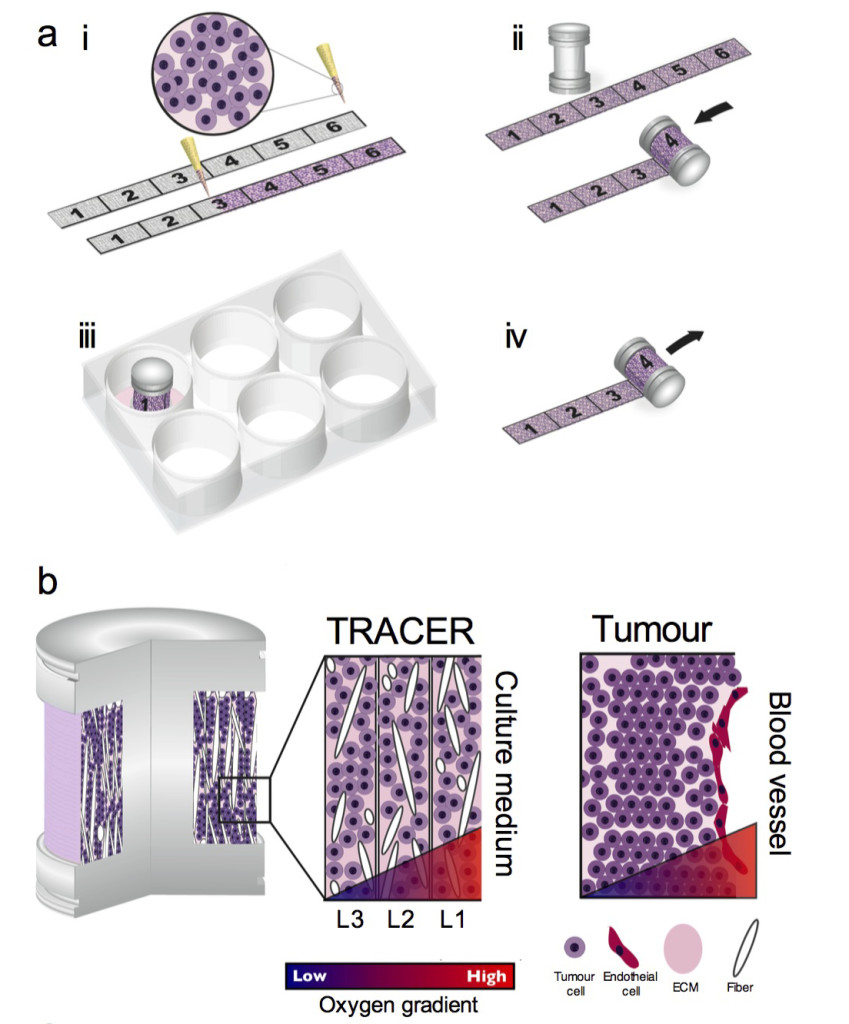
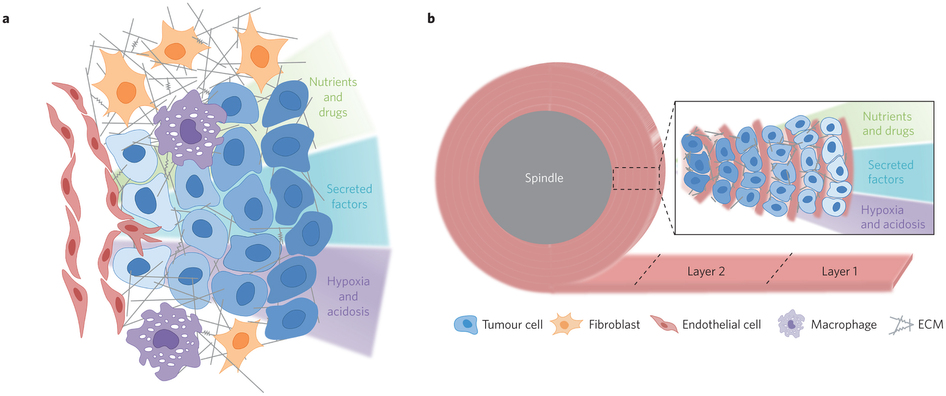
Concept of the TRACER: (i) cells are infiltrated into scaffold to form a biocomposite strip, (ii) the biocomposite strip is rolled on to an oxygen impermeable core, (iii) the TRACER is cultured for desired time, (iv) the TRACER is unrolled for analysis and location of the cells in 3D determined by their position along the strip. b – Geometry comparison of TRACER and tumours. Oxygen and nutrient consumption through the layers of the TRACER is predicted to generate gradients mimicking those observed in tumours in vivo at progressively further distances from a blood vessel.